Laboratory Practice
Select publications that evaluate technical aspects of WGS, including analytical validity, quality metrics and best practices
Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: a retrospective diagnostic accuracy and prospective clinical validation study.
Ibanez K, Polke J, Hagelstrom RT, et al. Neurol. 2022 Mar;21(3):234-245. doi: 10.1016/S1474-4422(21)00462-2
A randomized controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants.
Kingsmore SF, Cakici JA, Clark MM. A J Hum Gen, 2019;105:1-15.
NSIGHT2 is a prospective, randomized-controlled trial comparing rapid whole genome sequencing (WGS) and rapid whole exome sequencing (WES) to infants <4 months of age within 96 hours of NICU/PICU admission or onset of features. Singleton analysis was performed for all samples with secondary trio reanalysis performed as reflex to negative results. Gravely ill infants underwent trio, ultra-rapid WGS. The combined diagnostic yield in this study was 23%. Yield was not significantly different between rapid WGS (20%) and rapid WES (19%); however, it was higher (24%) in ultra-rapid WGS. Median time to diagnosis was similar between rWGS and rWES (11.0 d vs. 11.2 d). urWGS and rWGS combined identified two times more pathogenic and likely pathogenic variants than WES. The authors conclude that the analytical performance of rWGS is superior to rWES supporting the use of rWGS as a first-tier test in the NICU/PICU setting.
A rigorous interlaboratory examination of the need to confirm next-generation sequencing-detected variants with an orthogonal method in vlinical genetic testing.
Lincoln SE, Truty R, Lin CF, Zook JM, Paul J, Ramey VH, Salit M, Rehm HL, Nussbaum RL, Lebo MS. J Mol Diagn. 2019 Mar;21(2):318-329. doi: 10.1016/j.jmoldx.2018.10.009.
Good laboratory practice for clinical next-generation sequencing informatics pipelines.
Gargis AS, Kalman L, Bick DP, da Silva C, Dimmock DP, Funke BH, Gowrisankar S, Hedge MR, Kulkarni S, Mason CE, Nagarajan R, Voelkerding KV, Worthey EA, Aziz N, Barnes J, Bennett SF, Bisht H, Church DM, Dimitrova Z, Gargis SR, Hafez N, Hambuch T, Hyland FCL, Luna RA, MacCannell D, Mann T, McCluskey MR, McDaniel TK, Ganova-Raeva LM, Rehm HL, Reid J, Campo DS, Resnick RB, Ridge PG, Salit ML, Skums P, Wong LJC, Zehnbauer BA, Lubin IM. Nat Biotechnol. 2015;33(7):689–693. doi:10.1038/nbt.3237.
Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children.
Mestek-Boukhibar l, Clement E, Jones WD, Drury S, Ocaka L, Gagunashvili A, Le Quesne Stabej P, Bacchelli C, Jani N, Rahman S, Jenkins L, Hurst JA, Bitner-Glindzicz M, Peters M, Beales PL, Williams HJ. J Med Genet. 2018; 55:721–728.
An open resource for accurately benchmarking small variant and reference calls.
Zook JM, McDaniel J, Olson ND, Wagner J, Parikh H, Heaton H, Irvine SA, Trigg L, Truty R, McLean CY, De La Vega FM, Xiao C, Sherry S, Salit M. Nat Biotechnol. 2019;37(5):561-566.
Best practices for benchmarking germline small-variant calls in human genomes.
Krusche P, Trigg L, Boutros PC, Mason CE, De La Vega FM, Moore BL, Gonzalez-Porta M, Eberle MA, Tezak Z, Lababidi S, Truty R, Asimenos G, Funke B, Fleharty M, Chapman BA, Salit M, Zook JM, Global Alliance for Genomics and Health Benchmarking Team. Nat Biotechnol. 2019 May;37(5):555-560. doi: 10.1038/s41587-019-0054-x.
Clinical Utility
Select publications evaluating changes in medical management as a result of WGS-derived test results
Effect of whole-genome sequencing on the clinical management of acutely ill infants with suspected genetic disease: A randomized clinical trial.
Krantz ID, Medne L, Weatherly JM, et al. JAMA Pediatr. 2021;175(12):1218-1226. doi:10.1001/jamapediatrics.2021.3496
In this randomized control trial, 354 critically ill infants received WGS and were randomized to receive results at 15 days (Early) or 60 days (Delayed) after enrollment. The most common indication for testing was the presence of multiple congenital anomalies (57%). A change in management was twice as likely (21% vs. 10%; p=0.009) with WGS vs. usual care testing. The most common being: subspecialty referrals, alternations in medication, surgery or other procedures. A two-fold increase in diagnostic yield was found with WGS compared to usual care testing. The authors conclude that for acutely ill infants in the intensive care unit, first-line WGS is associated with a significant increase in clinical management compared to usual care. These data support WGS adoption and implementation in this setting.
100,000 genomes pilot on rare-disease diagnosis in health care – preliminary report.
Smedly D, Smith KR, Martin A, et al (100,000 Genomes Project Pilot Investigators). N Engl J Med. 2021 Nov 11;385(20):1868-1880.doi: 10.1056/NEJMoa2035790.
In 4660 participants of all ages who underwent WGS, a new diagnosis was reported in 25% who had no previous diagnosis from standard of care testing. Diagnostic yield was highly variable based on indication. Of those who received a diagnosis, 14% were found in regions of the genome that would have been missed by other testing modalities. Immediate implications were reported in 25% of those who received a diagnosis. Genome sequencing in a national health care system uncovered a new diagnosis across a broad range of rare diseases.
Project baby bear: Rapid precision care incorporating rWGS in 5 California children’s hospitals demonstrates improved clinical outcomes and reduced costs of care.
Dimmock D, Caylor S, Waldman B, Benson W, Ashburner C, Carmichael JL, Carroll J, Cham E, Chowdhury S, Cleary J, D’Harlingue A, Doshi A, Ellsworth K, Galarreta CI, Hobbs C, Houtchens K, Hunt J, Joe P, Joseph M, Kaplan RH, Kingsmore SF, Knight J, Kochhar A, Kronick RG, Limon J, Martin M, Rauen KA, Schwarz A, Shankar SP, Spicer R, Rojas MA, Vargas-Shiraishi V, Wigby K, Zadeh N, Farnaes L. Am J Hum Genet. 2021 May 29S0002-9297(21)00192-0. doi: 10.1016/j.ajhg.2021.05.008. Epub ahead of print. PMID: 34089648.
Rapid WGS was performed on 184 critically ill infants with Medi-Cal who met study criteria. Outcomes were measured by healthcare provider questionnaires and modeled processes to estimate clinical management changes and cost savings to the hospital. A total of 74/184 (40%) infants received a diagnosis and 58 of 184 (32%) experience at least one change in medical care. Both positive and negative results led to changes in management and/or cost savings. Cost savings was calculated based on a smaller cohort of 31 infants whose length of stay was impacted by rWGS results. Considering the cost of rWGS for the entire cohort, the average cost savings was $2549-6294 per child. The authors concluded that rapid WGS can be deployed as a first-tier test in the NICU.
Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected diseases.
Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, Kingsmore SF. NPJ Genom Med. 2018 Jul 9;3:16. doi: 10.1038/s41525-018-0053-8.
This meta-analysis examined 37 studies comprising 20,068 children from January 2011 to August 2017. Overall, the diagnostic utility was 8.3 times greater with WES/WGS (36%/41%) compared to CMA (10%). 25.7% of diagnostic variants identified by WGS were not apparent by WES. WGS detected diagnostic variants beyond the scope of WES, including intronic SNVs, SNVs in noncoding RNA, small CNVs, and mitochondrial DNA mutations, as well as exonic SNVs under-covered by WES.
Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico.
Scocchia A, Wigby KM, Masser-Frye D, Del Campo M, Galarreta CI, Thorpe E, McEachern J, Robinson K, Gross A, ICSL Interpretation and Reporting Team, Ajay SS, Rajan V, Perry DL, Belmont JW, Bentley DR, Jones MC, Taft R. NPJ Genom Med. 2019;4:5.
In a resource limited setting, clinical WGS was provided at no-cost to 60 children with a mean age of 7.6 years who met testing criteria. Indications included 77% with suspected pattern of malformation and 23% with primary neurological presentation. The overall diagnostic yield was 68%. 41/60 had a genomic finding consistent with the phenotype. This included 76% of those referred for suspected malformations and 43% referred for primary neurological presentation. (p=0.0455). Post-test counseling was modified for both patient with and without a molecular diagnosis. The absence of diagnosis helpful in 6 of 19 cases without molecular diagnosis.
Leveraging rapid genome sequencing to alter care plans for pediatric patients in a community hospital setting in the United States.
Beuschel J, Geyer H, Rich M, et al. J Pediatr. 2021; 2395-239. doi: 0.1016/j.jpeds.2021.08.010.
Genome sequencing demonstrates high diagnostic yield in children with undiagnosed global developmental delay/intellectual disability: A prospective study.
Sun Y, Peng J, Liang D, et al. Hum Mutat. 2022 May;43(5):568-581.doi: 10.1002/humu.24347. Epub 2022 Mar 1.
A prospective study of parental perceptions of rapid whole-genome and -exome sequencing among seriously ill infants.
Cakici JA, Dimmock DP, Caylor SA, Gaughran M, Clarke C, Triplett C, Clark MM, Kingsmore SF, Bloss CS. Am J Hum Genet. 2020;107:953-962.
Diagnostic yield and treatment impact of whole-genome sequencing in paediatric neurological disorders.
Lee HF, Chi CS, Tsai CR. Dev Med Child Neurol. 2020: https://doi.org/10.1111/dmcn.14722
An RCT of rapid genomic sequencing among seriously ill infants results in high clinical utility, changes in management and low perceived harm.
Dimmock DP, Clark MM, Gaughran M, Cakici JA, Caylor SA, Clarke C, Feddock M, Chowdhury S, Salz L, Cheung C, Bird LM, Hobbs C, Wigby K, Farnaes L, Bloss CS, Kingsmore SF. Am J Hum Genet. 2020; 107(5):942-952.
Genome sequencing as a diagnostic test in children with unexplained medical complexity.
Costain G, Walker S, Marano M, Veenma D, Snell M, Curtis M, Luca S, Buera J, Arje D, Reuter MS, Thiruvahindrapuram B, Trost B, Sung WWL, Yuen RKC, Chitayat D, Mendoza-Londono R, Stavropoulos J, Scherer SW, Marshall CR, Cohn RD, Cohen E, Orkin J, Meyn MS, Hayeems RZ. JAMA Netw Open. 2020;3(9):e2018109.
The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants.
Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, Saunders CJ, Thiffault I, Miller NA, Zellmer L, Herd SM, Holmes AM, Batalov S, Veeraraghavan N, Smith LD, Dimmock DP, Leeder JS, Kingsmore SF. NPJ Genom Med. 2018 Feb 9;3:6. doi: 10.1038/s41525-018-0045-8.
Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine
Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, Nalpathamkalam T, Pellecchia G, Yuen RKC, Szego MJ, Hayeems RZ, Shaul RZ, Brudno M, Girdea M, Frey B, Alipanahi B, Ahmed S, Babul-Hirji R, Badilla Porras R, Carter MT, Chad L, Chaudhry A, Chitayat D, Jougheh Doust S, Cytrynbaum C, Dupuis L, Ejaz R, Fishman L, Guerin A, Hashemi B, Helal M, Hewson S, Inbar-Feigenberg M, Kannu P, Karp N, Kim RH, Kronick J, Liston E, MacDonald H, Mercimek-Mahmutoglu S, Mendoza-Londono R, Nasr E, Nimmo G, Parkinson N, Quercia N, Raiman J, Roifman M, Schulze A, Shugar A, Shuman C, Sinajon P, Siriwardena K, Weksberg R, Yoon G, Carew C, Erickson R, Leach RA, Klein R, Ray PN, Meyn MS, Scherer SW, Cohn RD, Marshall CR. NPJ Genomic Medicine, 2016; 1.
Rapid whole genome sequencing has clinical utility in the PICU
Sanford EF, Clark MM, Farnaes L, Williams MR, Perry JC, Inguli EG, Sweeney NM, Doshi A, Gold JJ, Briggs B, Bainbridge MN, Feddock M, Watkins K, Chowdhury S, Nahas SA, Dimmock DP, Kingsmore SF, Coufal NG. Rapid whole genome sequencing has clinical utility in children in the PICU. PediatrCritCare Med. 2019 Jun 19. doi: 10.1097/PCC.0000000000002056.
Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization.
Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, Cakici, JA, Benson W, Kaplan RH, Kronick R, Bainbridge MN, Friedman J, Gold JJ, Ding Y, Veeraraghavan N, Dimmock D, Kingsmore SF. NPJ Genom Med. 2018 Apr 4;3:10. doi: 10.1038/s41525- 018-0049-4.
Economic Utility
Select publications that evaluate the cost effectiveness of clinical WGS
Project baby bear: Rapid precision care incorporating rWGS in 5 California children’s hospitals demonstrates improved clinical outcomes and reduced costs of care.
Dimmock D, Caylor S, Waldman B, Benson W, Ashburner C, Carmichael JL, Carroll J, Cham E, Chowdhury S, Cleary J, D’Harlingue A, Doshi A, Ellsworth K, Galarreta CI, Hobbs C, Houtchens K, Hunt J, Joe P, Joseph M, Kaplan RH, Kingsmore SF, Knight J, Kochhar A, Kronick RG, Limon J, Martin M, Rauen KA, Schwarz A, Shankar SP, Spicer R, Rojas MA, Vargas-Shiraishi V, Wigby K, Zadeh N, Farnaes L. Am J Hum Genet. 2021 May 29S0002-9297(21)00192-0. doi: 10.1016/j.ajhg.2021.05.008. Epub ahead of print. PMID: 34089648.
Rapid WGS was performed on 184 critically ill infants with Medi-Cal who met study criteria. Outcomes were measured by healthcare provider questionnaires and modeled processes to estimate clinical management changes and cost savings to the hospital. A total of 74/184 (40%) infants received a diagnosis and 58 of 184 (32%) experience at least one change in medical care. Both positive and negative results led to changes in management and/or cost savings. Cost savings was calculated based on a smaller cohort of 31 infants whose length of stay was impacted by rWGS results. Considering the cost of rWGS for the entire cohort, the average cost savings was $2549-6294 per child. The authors concluded that rapid WGS can be deployed as a first-tier test in the NICU.
Estimating the burden and economic impact of pediatric genetic disease
Gonzaludo N, Belmont JW, Gainullin VG, Taft RJ. Genet Med. 2019. doi: 10.1038/s41436-019-0458-5. Epub ahead of print.
To estimate the economic burden of genetic disease in pediatric patients, the authors analyzed the 2012 KID database, a national all-payer database for children in US that includes 10% of uncomplicated births and 80% of complicated in-hospital births. Genetic disease-linked discharges were associated with higher healthcare utilization, including additional procedures (up to 4 more), longer length of stay (2-18 days) and higher total costs per discharge ($12,000-$77,000). Discharges with multiple genetic disease-linked diagnosis codes yield higher cost per discharge with an incremental increase of $13,999 per code (up to 7th code). Overall genetic disease-linked discharges account for a proportionately larger amount of the “national bill” vs. non genetic disease-linked.
Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization.
Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, Cakici, JA, Benson W, Kaplan RH, Kronick R, Bainbridge MN, Friedman J, Gold JJ, Ding Y, Veeraraghavan N, Dimmock D, Kingsmore SF. NPJ Genom Med. 2018 Apr 4;3:10. doi: 10.1038/s41525- 018-0049-4.
Cost-effectiveness of exome and genome sequencing for children with rare and undiagnosed conditions .
Lavelle TA, Feng X, Keisler M, et al. Genet Med.[published online ahead of print, 2022 Apr 8]. 2022;S1098-3600(22)00682-7. doi:10.1016/j.gim.2022.03.005
Diagnostic Utility
Relevant publications demonstrating the diagnostic potential of clinical WGS in selected patient populations
Molecular diagnostic yield of genome sequencing versus targeted gene panel testing in racially and ethnically diverse pediatric patients
Abdul-Husn NS, Marathe PN, Kelly NR, et al. Genet Med, 2023:in pre-print, https://doi.org/10.1016/j.gim.2023.100880
The NYCKidSeq project seeks to advance the use of genomic medicine for underserved children in New York City. The aim of this study was to evaluate the use of both genome sequencing (GS) and targeted gene panels (TGP) in diverse populations with suspected genetic disorders. Panel testing aligned with indications and included: cardiovascular, neurologic, and immunologic panels. Nearly half of the 642 probands were of Hispanic/Latino ancestry and 16% were Black/African American. The overall diagnostic yield of GS was nearly two times greater than TGP (16.5% vs. 8.1%; p<.001). Uncertain results were reported more commonly in probands of Black/African American ancestry vs. White/European (63.8% vs. 47.6%; P=.01). The authors conclude that GS improved diagnostic yield in Hispanic/Latino and White/European populations but not in those of Black/African American ancestry, highlighting the need for more genomic studies in non-European populations to improve variant classification capabilities. Additional publications from this cohort are in development.
Effect of Whole-Genome Sequencing on the Clinical Management of Acutely Ill Infants With Suspected Genetic Disease: A Randomized Clinical Trial.
Krantz ID, Medne L, Weatherly JM, et al. JAMA Pediatr. 2021;175(12):1218-1226. doi:10.1001/jamapediatrics.2021.3496
In this randomized control trial, 354 critically ill infants received WGS and were randomized to receive results at 15 days (Early) or 60 days (Delayed) after enrollment. The most common indication for testing was the presence of multiple congenital anomalies (57%). A change in management was twice as likely (21% vs. 10%; p=0.009) with WGS vs. usual care testing. The most common being: subspecialty referrals, alternations in medication, surgery or other procedures. A two-fold increase in diagnostic yield was found with WGS compared to usual care testing. The authors conclude that for acutely ill infants in the intensive care unit, first-line WGS is associated with a significant increase in clinical management compared to usual care. These data support WGS adoption and implementation in this setting.
100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care – Preliminary Report.
Smedly D, Smith KR, Martin A, et al (100,000 Genomes Project Pilot Investigators). N Engl J Med. 2021 Nov 11;385(20):1868-1880.doi: 10.1056/NEJMoa2035790.
In 4660 participants of all ages who underwent WGS, a new diagnosis was reported in 25% who had no previous diagnosis from standard of care testing. Diagnostic yield was highly variable based on indication. Of those who received a diagnosis, 14% were found in regions of the genome that would have been missed by other testing modalities. Immediate implications were reported in 25% of those who received a diagnosis. Genome sequencing in a national health care system uncovered a new diagnosis across a broad range of rare diseases.
Successful application of genome sequencing in a diagnostic setting: 1007 index cases from a clinically heterogeneous cohort.
Bertoli-Avella AM, Beetz C, Ameziane N, Rocha ME, Guatibonza P, Pereira C, Calvo M, Herrera-Ordonez N, Segura-Castel M, Diego-Alvarez D, Zawada M, Kandaswamy KK, Werber M, Paknia O, Zielske S, Ugrinovski D, Warnack G, Kampe K, Iurașcu MI, Cozma C, Vogel F, Alhashem A, Hertecant J, Al-Shamsi AM, Alswaid AF, Eyaid W, Al Mutairi F, Alfares A, Albalwi MA, Alfadhel M, Al-Sannaa NA, Reardon W, Alanay Y, Rolfs A, Bauer P. Eur J Hum Genet. 2021 Jan;29(1):141-153. doi: 10.1038/s41431-020-00713-9. Epub 2020 Aug 28. PMID: 32860008; PMCID: PMC7852664.
This study analyzed genome sequencing (GS) data from 1007 consecutive cases with a broad spectrum of clinical presentations with onset of symptoms ranging from prenatal to 59 years of age. The overall diagnostic yield was 21.1%. When excluding patients with prior exome sequencing (i.e. GS was performed as a first-tier test) , the diagnostic yield was 24.7%. In 358 cases with a prior inconclusive or negative ES, an incremental diagnostic yield of 14.5% (n=52) was reported; 11 out of 52 were due to the technical superiority of GS over ES. An additional 15.1% (n=54) of cases with a prior inconclusive ES received a VUS that may or may not explain the phenotype. The authors conclude that GS should be either a standard second-line, or even first-line stand-alone test.
Genome sequencing as a diagnostic test in children with unexplained medical complexity.
Costain G, Walker S, Marano M, Veenma D, Snell M, Curtis M, Luca S, Buera J, Arje D, Reuter MS, Thiruvahindrapuram B, Trost B, Sung WWL, Yuen RKC, Chitayat D, Mendoza-Londono R, Stavropoulos J, Scherer SW, Marshall CR, Cohn RD, Cohen E, Orkin J, Meyn MS, Hayeems RZ. JAMA Netw Open. 2020;3(9):e2018109.
This prospective study evaluated the analytical and clinical validity of whole genome sequencing (WGS) as a first-tier test compared to conventional genetic testing in a cohort of 49 children with medical complexity. The median number of conventional genetic tests was four per proband (1-13) including chromosomal microarray (n=48) and WES (n=33). For analytical validity evaluation, WGS at 36X detected 100% of variants (124). The overall diagnostic yield was 30.6% (15/49) and three new genetic conditions were discovered. Clinical implications were noted in 12 patients, including an immediate medical management change in 7. The authors conclude that WGS has a role as a first-tier test in children with medical complexity.
Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients.
Stranneheim H, Lagerstedt-Robinson K, Magnusson M, et al. Genome Med. 2021; 13(40): https://doi.org/10.1186/s13073-021-00855-5.
Whole genome sequencing (WGS) was performed in 3219 patients over a four-year period. Initial analysis was completed using disease-specific panels or an OMIM morbid gene panel based on phenotype. In the absence of a molecular finding, additional analysis including a research WGS, was offered. The overall diagnostic yield was 40% and variants were identified in 754 different genes. Variant analysis evolved over the course of the study to include all variant types currently available via WGS. The authors state that clinical WGS has turned out to be a true game changer in the rare disease space.
A randomized controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants.
Kingsmore SF, Cakici JA, Clark MM, Gaughran M, Feddock M, Batalov S, Bainbridge MN, Carroll J, Caylor SA, Clarke C, Ding Y, ellsworth K, Farnaes L, Hildreth A, Hobbs C, James K, King CI, Lenberg J, NahasS, Prince L, Reyes I, Salz L, Sanford E, Schols P, Sweeney N, Tokita M, Veeraraghavan N, Watkins K, Wigby K, Wong T, Chowdhury S, Wright MS, Dimmock D, RCIGM Investigators. A J Hum Gen, 2019;105:1-15.
NSIGHT2 is a prospective, randomized-controlled trial comparing rapid whole genome sequencing (WGS) and rapid whole exome sequencing (WES) to infants <4 months of age within 96 hours of NICU/PICU admission or onset of features. Singleton analysis was performed for all samples with secondary trio reanalysis performed as reflex to negative results. Gravely ill infants underwent trio, ultra-rapid WGS. The combined diagnostic yield in this study was 23%. Yield was not significantly different between rapid WGS (20%) and rapid WES (19%); however, it was higher (24%) in ultra-rapid WGS. Median time to diagnosis was similar between rWGS and rWES (11.0 d vs. 11.2 d). urWGS and rWGS combined identified two times more pathogenic and likely pathogenic variants than WES. The authors conclude that the analytical performance of rWGS is superior to rWES supporting the use of rWGS as a first-tier test in the NICU/PICU setting.
Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test
Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini M, Thiruvahindrapuram B, Merico D, Jobling R, Nalpathamkalam T, Pellecchia G, Sung WWL, Wang Z, Bikangaga P, Boelman C, Carter MT, Cordeiro D, Cytrynbaum C, Dell SD, Dhir P, Dowling JJ, Heon E, Hewson S, Hiraki L, Inbar-Feigenberg M, Klatt R, Kronick J, Laxer RM, Licht C, MacDonald H, Mercimek-Andrews S, Mendoza-Londono R, Piscione T, Schneider R, Schulze A, Silverman E, Siriwardena K, Snead OC, Sondheimer N, Sutherland J, Vincent A, Wasserman JD, Weksberg R, Shuman C, Carew C, Szego MJ, Hayeems RZ, Basran R, Stavropoulos DJ, Ray PN, Bowdin S, Meyn MS, Cohn RD, Scherer SW, Marshall CR. Genet Med. 2017; Aug 3. doi: 10.1038/gim.2017.119.
In this prospective comparison of WGS to standard clinical testing (including NGS panels) in 103 pediatric patients with diverse phenotypes, the authors concluded that WGS is superior given its higher diagnostic yield. WGS confirmed significantly more diagnoses than conventional testing (41% vs 24%; P =0.01). All copy number variants reported by chromosomal microarray were detected by WGS and WGS offered more complete coverage of disease associated genes compared to WES.
Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico.
Scocchia A, Wigby KM, Masser-Frye D, Del Campo M, Galarreta CI, Thorpe E, McEachern J, Robinson K, Gross A, ICSL Interpretation and Reporting Team, Ajay SS, Rajan V, Perry DL, Belmont JW, Bentley DR, Jones MC, Taft R. NPJ Genom Med. 2019;4:5.
In a resource limited setting, clinical WGS was provided at no-cost to 60 children with a mean age of 7.6 years who met testing criteria. Indications included 77% with suspected pattern of malformation and 23% with primary neurological presentation. The overall diagnostic yield was 68%. 41/60 had a genomic finding consistent with the phenotype. This included 76% of those referred for suspected malformations and 43% referred for primary neurological presentation. (p=0.0455). Post-test counseling was modified for both patient with and without a molecular diagnosis. The absence of diagnosis helpful in 6 of 19 cases without molecular diagnosis.
From cytogenetics to cytogenomics: whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability
Anna Lindstrand, Jesper Eisfeldt, Maria Pettersson, Claudia M. B. Carvalho, Malin Kvarnung, Giedre Grigelioniene, Britt-Marie Anderlid, Olof Bjerin, Peter Gustavsson, Anna Hammarsjö, Patrik Georgii-Hemming, Erik Iwarsson, Maria Johansson-Soller, Kristina Lagerstedt-Robinson, Agne Lieden, Måns Magnusson, Marcel Martin, Helena Malmgren, Magnus Nordenskjöld, Ameli Norling, Ellika Sahlin, Henrik Stranneheim, Emma Tham, Josephine Wincent, Sofia Ygberg, Anna Wedell, Valtteri Wirta, Ann Nordgren, Johanna Lundin and Daniel Nilsson. Genome Med. 2019; 11: 68. Published online 2019 Nov 7. doi: 10.1186/s13073-019-0675-1
Using short-read whole genome sequencing (WGS), the authors evaluated three cohorts to determine in which WGS would be an ideal first-tier diagnostic test. The cohorts included: a cohort with validated copy number variants (CNVs) (cohort 1, n=68), individuals referred for monogenic multi-gene panels (cohort 2, n=156), and 100 prospective, consecutive cases referred to chromosomal microarray (CMA). CMA was performed using a custom oligonucleotide microarray with a median probe spacing of approximately 18kb and WGS performed using PCR-free, paired-end sequencing at 30X. Overall 27% of individuals harbored clinically relevant genetic variants by WGS compared to 12% by CMA. The authors concluded that study showed the power of WGS as a first-tier diagnostic test to detect a variety of CNVs and SNVs as well as single tandem repeats, regions of heterozygosity and chromosomal rearrangements.
Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine
Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, Nalpathamkalam T, Pellecchia G, Yuen RKC, Szego MJ, Hayeems RZ, Shaul RZ, Brudno M, Girdea M, Frey B, Alipanahi B, Ahmed S, Babul-Hirji R, Badilla Porras R, Carter MT, Chad L, Chaudhry A, Chitayat D, Jougheh Doust S, Cytrynbaum C, Dupuis L, Ejaz R, Fishman L, Guerin A, Hashemi B, Helal M, Hewson S, Inbar-Feigenberg M, Kannu P, Karp N, Kim RH, Kronick J, Liston E, MacDonald H, Mercimek-Mahmutoglu S, Mendoza-Londono R, Nasr E, Nimmo G, Parkinson N, Quercia N, Raiman J, Roifman M, Schulze A, Shugar A, Shuman C, Sinajon P, Siriwardena K, Weksberg R, Yoon G, Carew C, Erickson R, Leach RA, Klein R, Ray PN, Meyn MS, Scherer SW, Cohn RD, Marshall CR. NPJ Genomic Medicine, 2016; 1.
Genome sequencing demonstrates high diagnostic yield in children with undiagnosed global developmental delay/intellectual disability: A prospective study.
Sun Y, Peng J, Liang D, et al. Hum Mutat. 2022 May;43(5):568-581.doi: 10.1002/humu.24347. Epub 2022 Mar 1.
Genome sequencing as a first-line diagnostic test for hospitalized infants.
Bowling KM, Thompson ML, Finnila CR, et al. Genet Med. 2022; 24(4): 851-861.
Diagnostic yield and treatment impact of whole-genome sequencing in paediatric neurological disorders.
Lee HF, Chi CS, Tsai CR. Dev Med Child Neurol. 2020: https://doi.org/10.1111/dmcn.14722
The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants.
Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, Saunders CJ, Thiffault I, Miller NA, Zellmer L, Herd SM, Holmes AM, Batalov S, Veeraraghavan N, Smith LD, Dimmock DP, Leeder JS, Kingsmore SF. NPJ Genom Med. 2018 Feb 9;3:6. doi: 10.1038/s41525-018-0045-8.
Interpretation of genomic sequencing results in healthy and ill newborns: Results from the BabySeq Project
Ceyhan-Birsoy O, Murry JB, Machini K, Lebo MS, Yu TW, Fayer S, Genetti CA, Schwartz TS, Agrawal PB, Parad RB, Holm IA, McGuire AL, Green RB, Rehm HL, Beggs AH, The BabySeq Project Team.. Am J Hum Genet. 2019; 104(1): 76–93. doi: 10.1016/j.ajhg.2018.11.016
Reviews
Commentaries and scoping literature reviews that discuss rare disease, genomic medicine and general NGS considerations
Meta-analysis of the diagnostic and clinical utility of exome and genome sequencing in pediatric and adult patients with rare disease across diverse populations
Chung CCY, Hue SPY, Ng NYT et al. Genet Med. 2023; https://doi.org/10.1016/j.gim.2023.100896
The aim of this meta-analysis was to compare the diagnostic and clinical utility of exome sequencing (ES) versus genome sequencing (GS) in patients with rare disease including papers published from 2012 to 2021. A total of 161 studies from 31 countries/regions reporting on 50,412 probands were evaluated after initial review. Half of the studies included exclusively pediatric probands, 43% were mixed pediatric and adult probands and 3% were exclusively adult. The remaining were unknown. The pooled diagnostic yield did not find a significant difference between ES and GS (38% vs. 34%; P=.01); however, the yield was higher in pediatric probands compared to adult probands. Clinical utility was reported in 62 publications noting that the proportion of patients experiencing a change in management was significantly greater following GS than ES (61% vs. 48%; p=0.07). Differences in diagnostic yield between ES and GS were also assessed with respect to trio vs. proband-only testing, clinical indication, rapid vs. non-rapid testing and number of variants of uncertain significance. To date, this is the most comprehensive comparison of diagnostic and clinical utility between ES and GS. Despite similar diagnostic yields between the two modalities, clinical utility following GS is significantly greater than that of ES.
Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability.
Malinowski J, Miller DT, Demmer L, Gannon J, Pereira EM, Schroeder MC, Scheuner MT, Chun-Hui Tsai A, Hickey SE, Shen J on behalf of the ACMG Professional Practice and Guidelines Committee.
This review compiled primary literature from 2007 to 2019 to evaluate the utility of exome/ genome sequencing in patients with congenital anomalies, developmental delay or intellectual disability (CA/DD/ID). After review of 7178 publications, 167 met criteria for evaluation including 36 with >20 patients. For the studies with larger sample sizes, utility was evaluated following ES and/or GS (ES= 27; GS = 7; ES/GS = 2) with a range in sample size from 22-278. Ninety-five percent of studies reported a change in clinical management including changes in medication, procedures, specialist referrals, redirection of care and clinical trial opportunities. More than half of patients experienced a reported clinical impact related to a ES/GS diagnosis and more than half of studies with >20 patients identified an impact on family planning or reproductive decision-making. There was a paucity of data describing improved morbidity and mortality; however, there was sufficient, indirect evidence of the clinical and personal utility of exome/genome sequencing in patients with CA/DD/ID. The committee concluded that there is a need for a more formal framework to evaluate outcomes for ES/GS. This review will be used to guide the development of an evidence-based guideline.
Estimating cumulative point prevalence of rare disease: analysis of the Orphanet database.
Nguengang Wakap S, Lambert DM, Orly A, Rodwell C, Gueydan C, Lanneau V, Murphy D, Le Cam Y, Rath A.. Europ J Hum Genet. 2019;https://doi.org/10.1038/s41431-019-0508-0
Nguengang Wakap et al. analyzed epidemiological data in the Orphanet database to determine a cumulative point prevalence of rare disease. Analysis included information in database as of October 1, 2018. Based on all cases in the database, the authors estimate that RD affects at least 3.5-5.9% of the global population and approximately 72% of RD are reported to have a genetic etiology. This publication limits its calculations to only unique clinical disorders, therefore, removing the possibility of duplicate counting and overestimation.
Ontario Health (Quality). Genome-Wide Sequencing for Unexplained Developmental Disabilities or Multiple Congenital Anomalies: A Health Technology Assessment.
Ont Health Technol Assess Ser. 2020 Mar 6;20(11):1-178. PMID: 32194879; PMCID: PMC7080457.
Genome sequencing and implications for rare disorders.
Posey JE. Orphanet Journal of Rare Diseases. 2019; 14:153. https://doi.org/10.1186/s13023-019-1127-0
Genomic medicine for undiagnosed diseases
Wise AL, Manolio TA, Mesah GA, Peterson JF, Roden DM, Tamburro C, Williams MS, Green ED.. Lancet, 2019; 394: 533-40.
Case for genome sequencing in infants and children with rare, and undiagnosed genetic disease.
Bick D, Jones, M, Taylor SL, Taft RJ, Belmont J. J Med Genet 2019;0:1–9. doi:10.1136/jmedgenet-2019-106111.
The clinical utility of exome and genome sequencing across clinical indications: a systematic evidence review.
Hum Genet.2021 Oct;140(10):1403-1416.doi: 10.1007/s00439-021-02331-x. Epub 2021 Aug 8. Schick S, Mighton C, Uleryk E, Pechlivanoglou P, Bombard Y.
Societal Guidelines
Current guidelines from key medical societies
Indications for WES/WGS1,27
ACMG published an evidence-based guideline in 2021 which strongly recommends exome and genome sequencing as a first- or second-tier test in:
- Patients with one or more congenital anomalies prior to 1 year of age
- Patients with intellectual disability/developmental delay prior to 18 years of age
In addition, ACMG has published recommendations to consider exome or genome sequencing in a phenotypically affected individual when:
- Phenotype or family history data strongly implicate a genetic etiology, but the phenotype does not correspond with a specific disorder for which a genetic test targeting a specific gene is available on a clinical basis.
- A patient presents with a defined genetic disorder that demonstrates a high degree of genetic heterogeneity, making WES or WGS analysis of multiple genes simultaneously a more practical approach.
- A likely genetic disorder but specific genetic tests available for that phenotype have failed to arrive at a diagnosis.
- When a fetus presents with a likely genetic disorder in which specific genetic tests, including targeted sequencing tests, available for that phenotype have failed to arrive at a diagnosis.
Tools and Methods for Variant Classification2,3
ACMG and others note that a careful review of the evidence for and against pathogenicity are important in variant classification. Use of sequence, population, and disease databases can be useful when classifying variants, however, clinical laboratories should be aware of potential limitations. In addition, laboratories are encouraged to contribute to variant databases and to form collaborations with clinicians.
Genomic Interpretation and Reporting2,3,4
ACMG recommends that genetic reports should be concise, easy to understand and contain all essential testing elements, supporting evidence for variants and follow up recommendations, if indicated. Careful review of the patient’s phenotype and the method of ascertainment are important in test interpretation.
Secondary Findings2,4-10,24,25,28
The ACMG has published lists of medically actionable genes recommended for return if a pathogenic variant is detected as a secondary finding in clinical genomic sequencing, based on conditions with the potential for intervention and improved outcomes if caught early. They also created a secondary findings working group to maintain the list of medically actionable secondary findings (currently 73 genes). Several other groups recommend that laboratories and clinics offering WGS/WES should have clear policies in place related to disclosure of secondary findings. Informed consent should indicate an option to opt-in or opt-out of receiving secondary findings and include a discussion about ramifications of either decision.
Clinical Data Sharing2,11-13
Clinical genomic data sharing is an important aspect of genomic sequencing that has been addressed by multiple societies, including ACMG, AMP, CCMG, ESHG and NSGC. Data-sharing is essential to reduce the number of variants of uncertain significance and to improve consistency and accuracy of variant interpretation.
Confirmatory Testing3,14-16
There are a range of opinions from ACMG, AMP and CAP about whether confirmation of all pathogenic or likely pathogenic sequence variants is recommended.
Variant Reevaluation and Reanalysis17
Requests for variant reanalysis can come from the patient, healthcare provider or laboratory. Clinical laboratories should have separate policies and protocols for initial variant classification, variant-level reclassification, and case-level reanalysis, which should be periodically reviewed and updated.
Technical Standards26
The American College of Medical Genetics and Genomics developed a standard for the development of next-generation sequencing tests for constitutional variants. The following points are covered in this publication: clinical use of testing, technology and processes, test development and validation, reporting standards, data management, ongoing quality management and proficiency testing. This document does not apply to cell-free DNA analysis, infections disease testing or RNA applications of NGS.
Rare Disease Statistics
Up to 2-6%
of the population worldwide is affected by a rare disease (RD).1-6
Over 7,000
rare diseases have been identified.1,3,6,7
Among rare diseases listed by Orphanet,
~80% are either exclusively genetic or have genetic subtypes.1-3, 8
Half of Rare Disease cases impact children
and 30% of children affected by a RD will not survive beyond the age of 5 years.3,6
The average diagnostic odyssey lasts approximately
5-7 years.
6,9,10
Considering the cost per diagnosis, serial genetic testing in which WGS is used as a last resort test, is
less effective than use as a first-line test.11
For critically ill infants with a Rare Disease, a rapid diagnosis can be critical for timely and appropriate medical intervention.
An early diagnosis can prevent a long, expensive diagnostic journey.12-15
Genetic Testing Approaches for Rare Disease Diagnosis
-
Current standard of care for rare disease may include single gene testing, multi-gene panel testing, chromosomal microarray (CMA) and/or whole-exome sequencing (WES).
-
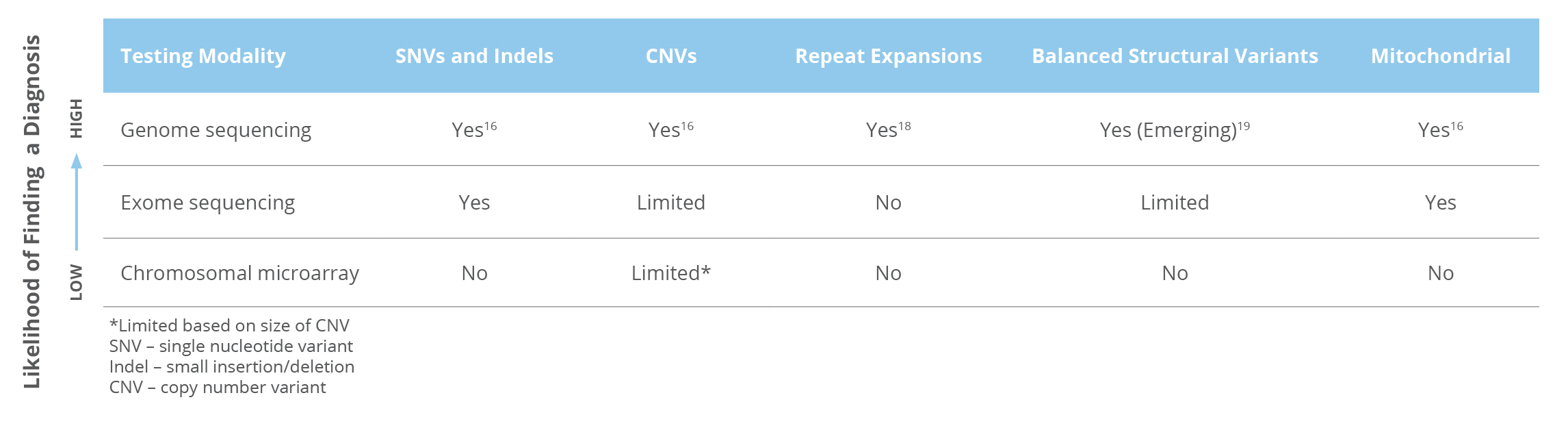
Whole genome sequencing (WGS) is the only test that can detect nearly all types of genetic variants. (Table 1) 16,17
Table 1
Guideline References
- American College of Medical Genetics and Genomics Board of Directors. Clinical utility of genetic and genomic services: A position statement of the American College of Medical Genetics and Genomics. Genet Med. 2015; 17(6): 505-507.
- Hegde M, Bale S, Bayrak-toydemir P, Gison J, Bone Jeng LJ, Joseph L, Laser J, Lubin IM, Miller CE, Ross LF, Rothberg PG, Tanner AK, VItazka P, Mao R. Reporting Incidental Findings in Genomic Scale Clinical Sequencing d A Clinical Laboratory Perspective A Report of the Association for Molecular Pathology. J Mol Diagnostics. 2015;17(2):107-117. doi:10.1016/j.jmoldx.2014.10.004.
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hedge M, Lyon E, Spector E, Voelkerding K, Rehm HL, on behalf of the ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424.
- Matthijs G, Souche E,, Alders M., Corveleyn A, Eck S, Feenstra I, Race V, Sistermans E, Sturm M, Weiss M, Yntema H, Bakker E, Scheffer H, Bauer P. Guidelines for diagnostic next-generation sequencing. Euro J Hum Genet. 2016;24:2–5.
- American College of Medical Genetics and Genomics (ACMG). Policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med. 2015;17(1):68-69.
- American College of Medical Genetics and Genomics. Points to consider in the clinical application of Genomic Sequencing. a position statement of the American college of medical genetics and genomics. May 2012. epub.
- Boycott K, Hartley T, Adam S, Bernier F, Chong K, Fernandez BA, Friedman JM, Geraghty MT, Hume S, Knoppers BM, Laberge AM, Majewski J, Mendoza-Londono R, Meyn MS, Michaud JL, Nelson TN, Richer J, Sadikovic B, Skidmore DL, Stockley T, Taylor S, van Karnebeek C, Zawati MH, Lauzon J, Armour CM on behalf of the Canadian College of Medical Genetics. The clinical application of genome-wide sequencing for monogenic diseases in Canada: Position Statement of the Canadian College of Medical Geneticists. J Med Genet. 2015;52:431-437.
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire A, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565-574.
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Herman GE, Hufnagel SB, Klein TE, Korf BR, McKelvey KD, Ormond KE, Richards CS, Vlangos CN, Watson M, Martin CL, Miller DT on behalf of the ACMG Secondary Findings Maintenance Working Group. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249-255.
- National Society of Genetic Counselors. Position Statement: Incidental findings in genetic testing. April 2019.
- American College of Medical Genetics and Genomics Board of Directors. Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(7):721-722.
- Position Statement on Licensed Databases and Plans for the Global Sharing of Variant Data. 2015. Accessed February 2019.
- National Society of Genetic Counselors. Clinical data sharing. Position Statement. April 2015.
- Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, Grody WW, Hedge MR, Hoeltge GA, Leonard DGB, Merker JD, Nagarajan R, Palicki LA, Robetorye RS, Schrijver I, Weck KE, Voelkerding KV. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2014;doi;10.5858/arpa.2014-0250-CP.
- Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hedge MR. Lyon E, from the Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733-47.
- Schrijver I, Aziz N, Farkas DH, Furtado M, Ferreira Gonzalez A, Greiner TC, Grody WW, Hambuch T, Kalman L, Kant JA, Klein RD, Leonard DGB, Lubin IM, Mao R, Nagan N, Pratt VM, Sobel ME, Voelkerding KV, Gibson JS. Opportunities and challenges associated with clinical diagnostic genome sequencing: A report of the association for molecular pathology. J Mol Diagn. 2012; 14(6): 525-540.
- Deignan JL, Chung WK, Kearney HM, Monaghan KG, Rehder CW, Chao EC, on behalf of the ACMG Laboratory Quality Assurance Committee. Points to consider in the reevaluation and reanalysis of genomic test results: a statement of the American College of Medical Genetics and Genomics (ACMG). On behalf of the ACMG Laboratory Quality Assurance Committee. Genet Med. 2019;
- American College of Medical Genetics and Genomics Board of Directors. The use of ACMG secondary findings recommendations for general population screening: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2019; https://doi.org/10.1038/s41436-019-0502-5.
- American College of Medical Genetics and Genomics Board of Directors. Points to consider for informed consent for genome/exome sequencing. Genet Med. 2013; 15(9):748-749.
- Bush, LW, Beck AE, Biesecker LG, Evans JP, Hamosh A, Holm IA, Martin CL, Richards CS, Rehm HL. Professional responsibilities regarding the provision, publication and dissemination of patietn phenotypes in the context of clinical genetic and genomic testing: Points to consider – a statement of the Amercian College of Medical Genetics and Genomics (ACMG). Genet Med. 2018.
- Gibson W, Stavropoulos J, Sinasac D, McCready E, Mahmutoglu S, Nelson TN. Canadian College of Medical Genetics Laboratory Practice Committee. CCMG statement on germline variant classification. 2017.
- Rooney Riggs E, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, Raca G, Ritter DI, South ST, Thorland EC, Pineda-Alvarez D, Aradhya S, Martin CL. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen).Genet Med. 2019: https://www.nature.com/articles/s41436-019-0686-8
- van El CG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV, Howard HC, Cambon-Thomsen A, Knoppers BM, Meijers-Heijboer H, Scheffer H, Tranebjaerg L, Dondorp W, de Wert GMWR on behalf of the ESHG Public and Professional Policy Committee. Whole genome sequencing in health care. Recommendations of the European Society of Human Genetics. Euro J Hum Genet. 2013;21:580-584.
- Miller DT, Lee K, Gordon AS, Amendola LM, Adelman K, Bale SJ, Chung WK, Gollob MH, Harrison SM, Herman GE, Hershberger RE, Klein TE, McKelvey K, Richards CS, Vlangos CN, Stewart DR, Watson MS, Martin CL & ACMG Secondary Findings Working Group. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med(2021). https://doi.org/10.1038/s41436-021-01171-4
- Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, Stewart DR, Amendola LM, Adelman K, Bale SJ, Gollob MH, Harrison SM, Hershberger RE, McKelvey K, Richards CS, Vlangos CN, Watson MS, Martin CL & ACMG Secondary Findings Working Group. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med (2021). https://doi.org/10.1038/s41436-021-01172-3
- Rehder C, Bean L.J.H., Bick D,Chao E, Chung W, Das S, O’Daniel J, Rehm H, Shasi V, Vincent LM, ACMG Laboratory Quality Assurance Committee. Next-generation sequencing for constitutional variants in the clinical laboratory, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med (2021). https://doi.org/10.1038/s41436-021-01139-4
- Manickam, K., McClain, Demmer LA, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG).. Genet Med (2021) https://doi.org/10.1038/s41436-021-01242-6
- Miller DT, Lee K, Abul-Husn NS, et al. ACMG SF v3.2 list for reporting of secondary findings in clinical exome and genome sequencing: A policy
statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2023: https://www.gimjournal.org/article/S1098-3600(23)00879-1/fulltext
Contact Us
Learn more about The Medical Genome Initiative and get answers to your questions.

The Medical Genome Initiative ©2022. All Rights Reserved.
