Overview
Published Manuscript, May 2020
Marshall, C.R., Bick, D., Belmont, J.W. et al. The Medical Genome Initiative: moving whole-genome sequencing for rare disease diagnosis to the clinic. Genome Med. 2020; 12:48. https://doi.org/10.1186/s13073-020-00748-z
Clinical whole-genome sequencing (WGS) offers clear diagnostic benefits for patients with rare disease. However, there are barriers to its widespread adoption, including a lack of standards for clinical practice. The Medical Genome Initiative consortium was formed to provide practical guidance and support the development of standards for the use of clinical WGS.
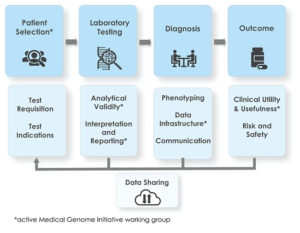
Analytical Validity
Published Manuscript, October 2020
Analytical validation of clinical whole genome sequencing for germline disease diagnostics: Best practices and performance standards. 📥
Marshall CR, Chowdhury S, Taft RJ, Lebo MS, Buchan JG, Harrison SM, Rowsey R, Klee EW, Liu P, Worthey EA, Jobanputra V, Dimmock D, Kearney HM, Bick D, Kulkarni S, Belmont JW, Stavropoulos DJ, Lennon NJ, on behalf of the Medical Genome Initiative.
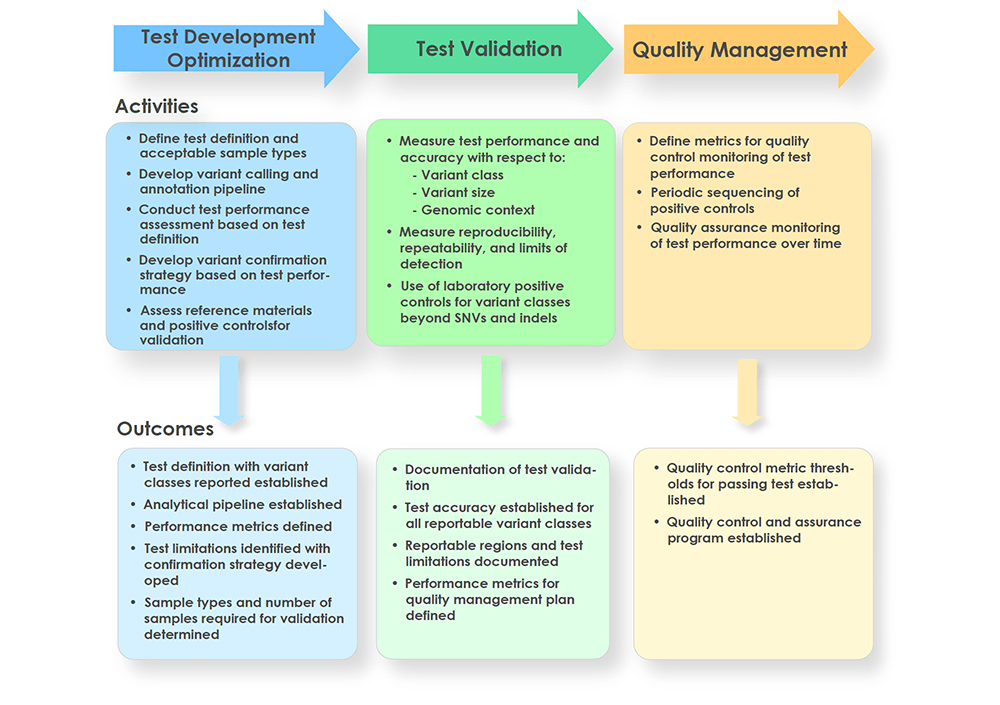
Whole-genome sequencing (WGS) has shown promise in becoming a first-tier diagnostic test for patients with rare genetic disorders, however, standards addressing the definition and deployment practice of a best-in-class test are lacking. To address these gaps, the Medical Genome Initiative, a consortium of leading health care and research organizations in the US and Canada, was formed to expand access to high quality clinical WGS by publishing best practices. Here, we present consensus recommendations on clinical WGS analytical validation with a focus on test development, upfront considerations for test design, test validation practices, and metrics to monitor test performance. This work also provides insight into the current state of WGS testing at each member institution, including the utilization of reference and other standards across sites. Importantly, members of this Initiative strongly believe that clinical WGS is an appropriate first-tier test for patients with rare genetic disorders and at minimum is ready to replace chromosomal microarray analysis and whole-exome sequencing. The recommendations presented here should reduce the burden on laboratories introducing WGS into clinical practice and support safe and effective WGS testing for diagnosis of germline disease.
Poster
Analytical validation of clinical whole genome sequencing for germline disease diagnostics: Best practices and performance standards. (PDF download) 📥
Marshall CR, Lennon NJ, Chowdhury S, Taft RJ, Stavropoulos DJ, Lebo MS, Harrison SM, Buchan JG, Liu P, Kulkarni S, Dimmock D, Belmont JW, Bick D, Worthey EA, Rowsey R, Klee EW, Kearney HM on behalf of the Medical Genome Initiative. Poster presented at: American Society of Human Genetics Annual Meeting; October, 2019; Houston, TX.
Clinical Utility
Published manuscript, December 2022
Clinical utility of genomic testing: A measurement toolkit 📥
Hayeems RZ, Dimmock DP, Bick DP, Belmont JW, Green RC, Lanpher B, Jobanputra V, Mendoza R, Kulkarni S, Grove ME, Taylor SL, Ashley E, on behalf of the Medical Genome Initiative.
Whole genome sequencing is emerging as the most robust strategy for achieving timely diagnoses in undiagnosed rare disease populations. Evidence of clinical utility and cost-effectiveness is required for WGS to be accepted into practice, commissioned in a health system, or receive reimbursement. Defining and measuring clinical utility is complex and context specific. The abstract addresses the need to develop a standardized framework and define measurement best practices to optimize the evidence base for decision makers and health care systems invested in providing high quality genome diagnostics.
Poster
Clinical utility of genomic testing: A measurement toolkit 📥
Hayeems RZ, Dimmock DP, Bick DP, Belmont JW, Green RC, Lanpher B, Jobanputra V, Mendoza R, Kulkarni S, Grove ME, Taylor SL, Ashley E, on behalf of the Medical Genome Initiative. Poster presented at: 2020 American College of Medical Genetics and Genomics Annual Clinical Genetics Meeting; May, 2020; Virtual
*This abstracted was also presented as poster no. P23.08.B at the European Society of Human Genetics meeting held virtually, June 6-9, 2020. The abstract has been accepted as an oral presentation at the European Conference on the Diffusion of Genomic Medicine in spring 2021 (dates to be determined).
Interpretation & Reporting
Published Manuscript, April 2022
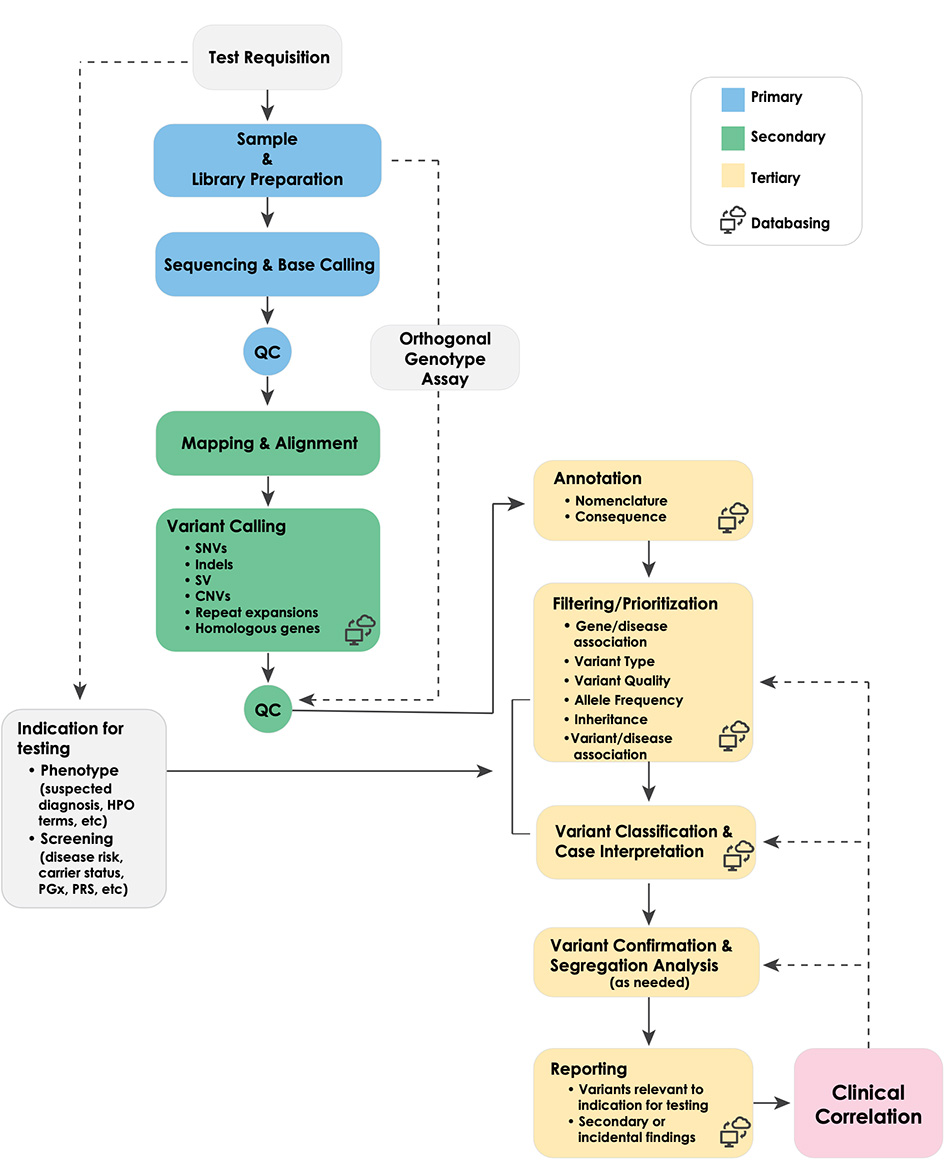
Best practices for the interpretation and reporting of clinical whole genome sequencing 📥
Christina A. Austin-Tse, Vaidehi Jobanputra, Denise L. Perry, David Bick, Ryan J. Taft, Eric Venner, Richard A. Gibbs, Ted Young, Sarah Barnett, John W. Belmont, Nicole Boczek, Shimul Chowdhury, Katarzyna A. Ellsworth, Saurav Guha, Shashikant Kulkarni, Cherisse Marcou, Linyan Meng, David R. Murdock, Atteeq U. Rehman, Elizabeth Spiteri, Amanda Thomas-Wilson, Hutton M. Kearney, Heidi L. Rehm and Medical Genome Initiative
Whole genome sequencing (WGS) shows promise as a first-tier diagnostic test for patients with rare genetic disorders. However, standards addressing the definition and deployment practice of a best-in-class test are lacking. To address these gaps, the Medical Genome Initiative, a consortium of leading health care and research organizations in the US and Canada, was formed to expand access to high quality clinical WGS by convening experts and publishing best practices. Here, we present best practice recommendations for the interpretation and reporting of clinical diagnostic WGS, including discussion of challenges and emerging approaches that will be critical to harness the full potential of this comprehensive test.
npj Genomic Medicine (2022)7:27 ; https://doi.org/10.1038/s41525-022-00295-z
Variants of Uncertain Significance
Published Manuscript, July 2023
The landscape of reported VUS in multi-gene panel and genomic testing: Time for a change📥
Rehm HL, Alaimo JT, Aradhya S, et al. The landscape of reported VUS in multi-gene panel and genomic testing: Time for a change. Genet Med. 2023. epub ahead of print. https://doi.org/10.1186/s13073-020-00748-z
Data was collected on 1.5 million inconclusive results with >1 variant of uncertain significance (VUS) from 19 clinical laboratories in the U.S. and Canada. Tests included multi-gene panel tests (MGP; 96.8%), exome sequencing (ES; 2.8%) and genome sequencing (GS; 0.4%). A significantly lower rate of inconclusive results due to VUS were reported by ES/GS compared to MGP (22.5% vs. 32.6%; p<0.0001). The use of trios led to significantly lower VUS rates (18.9% vs. 27.6%; p<0.0001). Additionally, the rate of VUS on MGP was directly correlated to panel size; the larger the panel, the higher the rate of VUS. The significantly higher rate of VUS from MGPs compared to ES/GS is best explained by current laboratory practices of reporting all VUS during diagnostic MGP compared to clinical correlation and parental data applied during ES/GS testing, The authors provide considerations for how laboratories can minimize the rate of reported VUS.
Posters
Genomic sequencing tests generate less uncertainty and higher diagnostic yield compared to multi-gene panel-based tests: Results of over 1.5 million tests 📥
Rehm HL, Alaimo JT, Aradhya S, et al. Poster presented at the American Society of Human
Genetics Annual Meeting; October, 2022; Los Angeles, CA
Patient Selection
Evidence Review and Considerations for the Use of First Line Genome Sequencing to Diagnose Rare Germline Disorders
Wigby KM, Brockman D, Costain G, Hale C, Taylor SL, Belmont J, Bick D, Dimmock D, Fernbach S, Greally J, Jobanputra V, Kulkarni S, Spiteri E, Taft RJ. npj Genom. Med. 9, 15 (2024). https://doi.org/10.1038/s41525-024-00396-x
An expert review panel was convened by the Medical Genome Initiative to perform a focused literature review followed by an exploratory meta-analysis using random effects model to generate a point estimate for diagnostic yield accounting for cohort size and number of diagnoses per cohort. Seventy-one studies met inclusion criteria. Using the point effects model, the diagnostic yield of genome sequencing (GS) across all studies was 34%. Clinical utility was reported in 32% of studies with a range of 20-100%. Based on the extensive review, the expert panel developed five recommendations about when GS should be considered. Future studies in the pediatric and adult populations may enable further refinement of these recommendations.
Commentaries
Jobanputra V, Schroeder B, Rehm H, Shen W, Spiteri E, Nakouzi G, Taylor S, Marshall CR, Meng L, Kingsmore SF, Ellsworth K, Ashley E, Taft RJ on behalf of the Medical Genome Initiative
Contact Us
Learn more about The Medical Genome Initiative and get answers to your questions.

The Medical Genome Initiative ©2023. All Rights Reserved.
